Gene Family Evolution
Gene duplications and the origin of Novelty
Genes duplications contribute to the origin of novel traits in may ways (e.g. increased gene dosage, acquisition of new protein functions or expression domains, etc). It has also been suggested nascent gene duplicates can lead to new functions through the evolution of novel competitive or cooperative interactions with each other, but empirical examples of such a process are lacking.
Our work on the FLOWERING LOCUS T (FT) gene family, which is critically involved in the control of flowering time and plant architecture, has identified a variant that allows us to test this hypothesis. During sunflower domestication, selection nearly fixed an allele of one sunflower FT copy that causes this altered protein to interfere with the function of a second copy (Blackman et al., 2010).
Dramatic changes in which tissues and in what environments different FT copies are expressed have also evolved in within and between species in the sunflower lineage. By characterizing the evolutionary timing and molecular mechanisms underlying this variation, our lab is working to gain additional insights into how gene duplications evolve new functions and contribute to diversity in phenotypic plasticity.
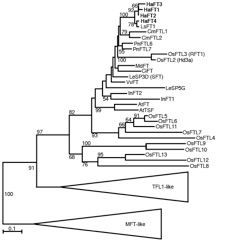

Phylogeny of FLOWERING LOCUS T homologs
Wild and domesticated alleles of a sunflower FT gene differ in their ability to rescue ft mutants when transformed into Arabidopsis thaliana