| Primer list (private) | |
| Pickle | |
Flowering plants generally begin life with vegetative growth. Given appropriate environmental conditions, plants flower when they mature. Theoretically, vegetative development can result from activation of a vegetative program or repression of the reproductive program. So far, evidence supporting the former is scarce; few vegetative-specific genes have been found in Arabidopsis. On the other hand, there is ample evidence that floral repression is sufficient for maintaining vegetative development.
Loss-of-function mutants in the EMBRYONIC FLOWER (EMF) genes, EMF1 and EMF2, skip vegetative growth and flower upon germination (Sung et al., 1992). Removal of the floral repressors results in early flowering and even the elimination of vegetative growth, consistent with the notion that the reproductive or flowering state is the default state in Arabidopsis.
EMF1 encodes a plant-specific nuclear protein, and EMF2, a Polycomb group (PcG) protein homolog. In plants and animals the PcG proteins form multimeric epigenetic silencer complexes that control diverse developmental pathways. Two functionally distinct PcG complexes that generate repressive chromatin states around target genes, Polycomb Repressive Complex (PRC) 2 and PRC1, exist in animals. PRC2, with four core proteins, Extra sex combs (Esc), P55, Enhancer of zeste (E(z)) and Suppressor of zeste (Su(z)12), can methylate histone H3 at lysine 27 through the E(z) SET domain. This methylation has been proposed to act as a molecular mark that recruits the PRC1, which contains Polycomb (Pc), Polyhomeotic (Ph), dRing1, and Psc as well as components of the basal transcription factor TFIID.
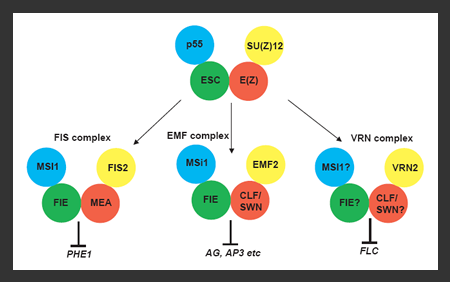
Fig. Arabidopsis Polycomb-group protein complexes. The core components of the Drosophila PRC2 complex are shown at top. In Arabidopsis, an equivalent ancestral complex is proposed to have
diversified into three similar complexes with at least partially discrete
functions. The colours indicate homology. (1) FIS2/MEA/FIE/MSI1 mediates Pheres1 repression of endosperm proliferation during gametophyte and endosperm development ; (2) EMF2/CLF/FIE/MSI1 represses the flower homeotic genes Agamous (AG), Apetalla 3 (AP3), and Pistallata (PI) during vegetative development; and (3) the VRN2 complex exercises epigenetic control of the vernalization response by repressing Flowering Locus C (FLC). EMF2 belongs to a small Arabidopsis gene family involved in PcG complexes that specify developmental processes through the repression of MADS-box genes.
Arabidopsis possesses three PRC2-like complexes (see Figure). The Arabidopsis Esc and p55 orthologs, Fertilization Independent Endosperm (FIE) and Multicopy Suppressor of IRA1(MSI1), respectively, probably are present in all three complexes. Arabidopsis has multiple copies of the Su(z)12 and E(z) homologs: EMF2, Fertilization Independent Endosperm2 (FIS2) and Vernalization2 (VRN2) are similar to Su(z)12, while, MEDEA (MEA), Curly Leaf (CLF), and Swinger (SWN) are E(z) homologs. The three complexes regulate at least three different processes via distinct MADS-box proteins (see figure). A modest increase in the PcG protein genes that form multi-protein complexes has expanded the capability and diversity of the regulatory network.
We are interested in elucidating the mechanism of EMF-mediated floral repression. A variety of genetic, molecular, functional genomic, biochemical and evolutionary approaches are employed to understand the function and evolution of the EMF genes.
Current Projects:
Function of EMF1.
Temporal and spatial requirement of EMF1.
In vivo interaction of the members of the EMF2 complex.
Role of DNA methylation in EMF-mediated floral repression.
Global gene expression pattern in mutants impaired in EMF protein complex.
Phylogenetic analysis of the VEF gene family.