A very large phage (red, left) as compared to normal phages infecting a bacterial cell. The UC Berkeley team of researchers hasn't yet photographed these large phages, so those depicted here resemble a common phage type called T4. Image courtesy of Jill Banfield lab.
Scientists have discovered hundreds of unusually large, bacteria-killing viruses with capabilities normally associated with living organisms, blurring the line between living microbes and viral machines.
These phages — short for bacteriophages, so-called because they “eat” bacteria — are of a size and complexity considered typical of life, carry numerous genes normally found in bacteria and use these genes against their bacterial hosts.
University of California, Berkeley, researchers and their collaborators found these huge phages by scouring a large database of DNA that they generated from nearly 30 different Earth environments, ranging from the guts of premature infants and pregnant women to a Tibetan hot spring, a South African bioreactor, hospital rooms, oceans, lakes and deep underground.
Altogether they identified 351 different huge phages, all with genomes four or more times larger than the average genomes of viruses that prey on single-celled bacteria.
Among these is the largest bacteriophage discovered to date: Its genome, 735,000 base-pairs long, is nearly 15 times larger than the average phage. This largest known phage genome is much larger than the genomes of many bacteria.
“We are exploring Earth’s microbiomes, and sometimes unexpected things turn up. These viruses of bacteria are a part of biology, of replicating entities, that we know very little about,” said Jill Banfield, a UC Berkeley professor of earth and planetary science and of environmental science, policy and management, and senior author of a paper about the findings appearing Feb 12 in the journal Nature. “These huge phages bridge the gap between non-living bacteriophages, on the one hand, and bacteria and Archaea. There definitely seem to be successful strategies of existence that are hybrids between what we think of as traditional viruses and traditional living organisms.”
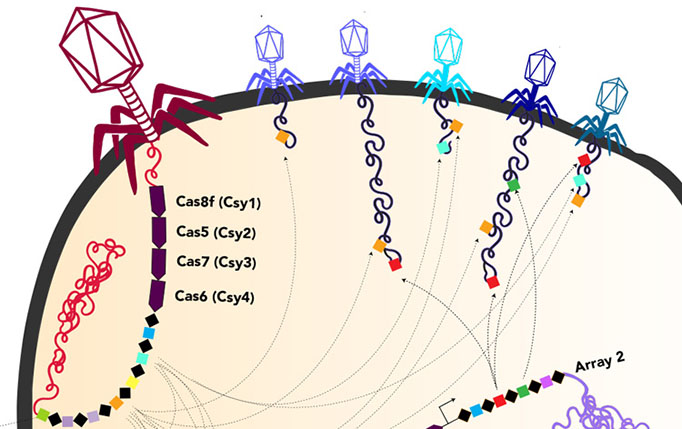
Ironically, within the DNA that these huge phages lug around are parts of the CRISPR system that bacteria use to fight viruses. It’s likely that once these phages inject their DNA into bacteria, the viral CRISPR system augments the CRISPR system of the host bacteria, probably mostly to target other viruses.
“It is fascinating how these phages have repurposed this system we thought of as bacterial or archaeal to use for their own benefit against their competition, to fuel warfare between these viruses,” said UC Berkeley graduate student Basem Al-Shayeb. Al-Shayeb and research associate Rohan Sachdeva are co-first authors of the Nature paper.
New Cas protein
One of the huge phages also is able to make a protein analogous to the Cas9 protein that is part of the revolutionary tool CRISPR-Cas9 that Jennifer Doudna of UC Berkeley and her European colleague, Emmanuelle Charpentier, adapted for gene-editing. Banfield’s team dubbed this tiny protein CasØ, because the Greek letter Ø, or phi, has traditionally been used to denote bacteriophage.
“In these huge phages, there is a lot of potential for finding new tools for genome engineering,” Sachdeva said. “A lot of the genes we found are unknown, they don’t have a putative function and may be a source of new proteins for industrial, medical or agricultural applications.”
Aside from providing new insight into the constant warfare between phages and bacteria, the new findings also have implications for human disease. Viruses, in general, carry genes between cells, including genes that confer resistance to antibiotics. And since phages occur wherever bacteria and Archaea live, including the human gut microbiome, they can carry damaging genes into the bacteria that colonize humans.
“Some diseases are caused indirectly by phages, because phages move around genes involved in pathogenesis and antibiotic resistance,” said Banfield, who is also director of microbial research at the Innovative Genomics Institute (IGI) and a CZ Biohub investigator. “And the larger the genome, the larger the capacity you have to move around those sorts of genes, and the higher the probability that you will be able to deliver undesirable genes to bacteria in human microbiomes.”
-The full article originally appeared and can be read on the Berkeley News website.
READ MORE:
